DNA methylation biomarkers are associated with diabetic kidney disease
More than nine million people worldwide have type 1 diabetes – a disease characterized by chronically elevated blood glucose levels caused by an immune-mediated destruction of the pancreatic insulin-producing cells. Despite treatment efforts to normalize blood glucose levels, the elevated blood glucose levels can damage the blood vessels e.g., in the kidney, causing diabetic kidney disease which can lead to kidney failure. The Finnish Diabetic Nephropathy Study, FinnDiane, is a nationwide study with over 5,000 participants with type 1 diabetes with the aim to identify genetic, environmental, and clinical risk factors leading to diabetic complications.
Epigenetic changes, such as DNA methylation, may contribute to the complex interaction between genes and the environment leading to disease. DNA methylation is a reversible chemical modification of DNA, commonly occurring in response to the environment, and changing the gene expression without altering the gene sequence itself. By studying blood DNA methylation levels at ~850 000 genomic sites in 1,304 individuals with type 1 diabetes, researchers in an international collaboration from the FinnDiane study and Queen’s University Belfast, UK, discovered 32 sites in the genome that were differentially methylated in individuals with diabetic kidney disease. More than half of these sites were situated in genes whose expression in kidneys was correlated to diabetic kidney disease or pathological changes in the kidneys. The research both discovered known genes for kidney disease, as well as revealing novel distinctive genes for diabetic kidney disease. In addition, the methylation levels at 21 genomic sites were able to predict the future risk of kidney failure in individuals with diabetes.
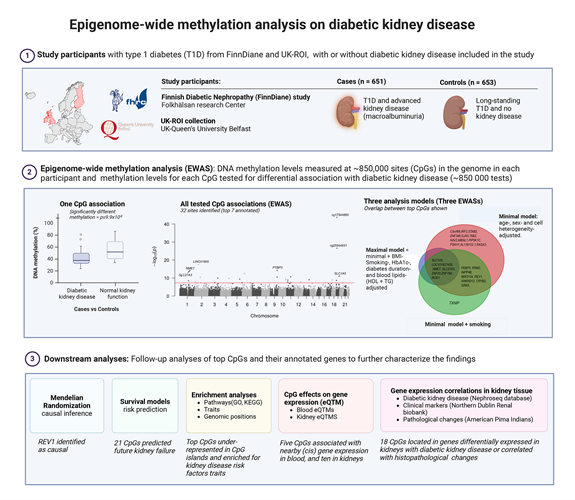
Graphical abstract. Created with BioRender.com.
Due to the reversibility of DNA methylation, the identified sites may even offer future targets for therapeutic intervention if determined causal. One such potential causal target for diabetic kidney disease was also identified within the REV1 gene by Mendelian Randomization analysis that utilized genetic information to infer causality.
Epigenome-wide meta-analysis identifies DNA methylation biomarkers associated with diabetic kidney disease
Smyth LJ*, Dahlström EH*, Syreeni A, Kerr K, Kilner J, Doyle R, Brennan E, Nair V, Fermin D, Nelson RG, Looker HC, Wooster C, Andrews D, Anderson K, McKay GJ, Cole JB, Salem RM, Conlon PJ, Kretzler M, Hirschhorn JN, Sadlier D, Godson C, Florez JC; GENIE consortium; Forsblom C, Maxwell AP, Groop PH, Sandholm N*, McKnight AJ*. Nature Communications.
